Archival of Staphopia
As of August 1st, 2023, the Staphopia resources have been placed in an archival or end-of-life state. The Staphopia project initially started in 2011 with an idea to process all the available Staphylococcus aureus genomes available at the time (~700), 12 years later there are now more than 120,000 S. aureus genomes available! During this time the field has changed significantly and many of the resources once available from only Staphopia are now available directly from public repositories. This evolution of the field has been fun to watch, and we hope you were able to put Staphopia to use in your research!
Can I still use the Staphopia analysis pipeline?
The Staphopia analysis pipline will no longer be supported, instead you should use Bactopia. Bactopia is the successor to Staphopia and is still being actively supported and developed. Even better Staphopia is built into Bactopia, so you can easily make use S. aureus specific analyses.
Can I access any assemblies from the original 44,000k genomes?
The assemblies from our original run are not available. However, you can use the Sample Metadata in combination with Grace Blackwell's 660k ENA assemblies to acquire assemblies based on the sample information. For example, you can use the Staphopia sample metadata table to identify which samples are ST8 and use the Experiment accession to pull the assemblies from Grace's set. Alternatively, you can use the Experiment accessions with Bactopia to reanalyze the samples with more current tools.
Below is text from the original website
Welcome To Staphopia
About Staphopia
Staphopia is a database focusing on the genomes of the bacterial pathogen Staphylococcus aureus. Life-threatening Methicillin-resistant S. aureus (MRSA) infections strike across our society, both in the community and hospital settings. In recent years whole genome shotgun sequencing of clinical isolates has become common. As of August 2017 there were over 44,000 publicly available S. aureus sequencing projects in EBI's ENA database. Staphopia aims to provide rapid analysis of these whole-genome datasets as well as useful S. aureus specific results, such as genotype, antibiotic resistance and virulence gene profiles.
Users can create an account to access a basic summary of results for each public sample through this site. Another option, is to access our web API with the use of our R package. For those interested in processing their own data, we have made both the Staphopia analysis pipeline and web application publicly available on GitHub (see below). We have also created a Docker image to simplify the installation process both locally and in the cloud. Also much of the Staphopia pipeline can be used a template for other organism-specific databases.
An Overview of Staphopia
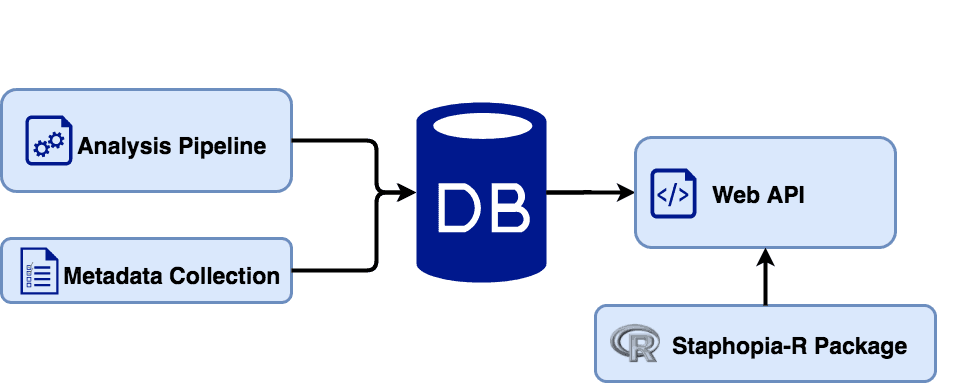
Archived Staphopia Resources
GitHub
- Staphopia-R
- An R package to query Staphopia's API
- Staphopia-AP
- Staphopia's analysis pipeline (Dockerized)
- Staphopia-Web
- Staphopia's Django powered web application
Acknowledgements
Staphopia was created by Robert Petit and Tim Read at Emory University School of Medicine in Atlanta Georgia, USA. We would like to thank Colleen Kraft, Tauqeer Alam, Santiago Castillo, Nikolay Braykov, Signe White, Michelle Su, Michael Frisch, Jim Hogan, King Jordan and Jim Heitner for help in putting together Staphopia. We acknowledge (and are grateful) for data from these sources: EBI's ENA, MIGS/MINS, S. aureus MLST, ARDB, StaphVar, and SCCmec.
Thanks also to those researchers and institutions who have contributed published and unpublished sequence public information in to the SRA databases.
Funding
Funding for this study from Emory University, Amazon AWS in Education Grant Program, and NIH grants AI091827 and AI121860. The Seven Bridges NCI Cancer Genomics Cloud pilot was supported in part by the funds from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN261201400008C.